Single Nucleotide Polymorphisms (SNPs) Are NOT the Only Type of Genetic Variants
Polymorphisms and genetic variants are collective terms that include more than just SNPs. For example, you have the following:
- Variation in the number of gene copies
- Repetitive sequence variations
- Addition and subtraction of DNA sequence to the gene
- Deletion of genes

Caption: different types of genetic variants in the human genome1
SNPs are the smallest genetic variants because they only involve one base pair. In the human genome, SNPs are found every 1000 base pairs. Eighty percent of all known human variants are SNPs, which may account for the majority of trait differences from person to person.2 However, SNPs are not the only type of genetic variations we have.
Keep in mind that currently, the only technology used by 23andme, Geno, and Ancestry are SNP arrays. They only detect SNPs. Therefore, it’s possible to have other larger and more impactful genetic variants than the SNPs you have or lack. SNPs are hardly the be-all and end-all of genetics.
How SNPs Got Associated With Traits and Diseases
Traditional geneticists (like me) identify genes responsible for a trait by mutating the genes and observing the resulting traits. Geneticists also “cross” or mate pairs of animals with the mutations of interest to understand how the mutations transmit through the generations and show up in the offspring. Throughout my academic career, I studied genetics in model organisms, including budding yeasts, flies, and worms.
In contrast, human geneticists have their hands tied because it’s neither ethical nor practical to carry out such genetic experiments in humans — they can only study existing humans. And because your genome has more than 20,000 genes and 3 Giga-base pairs (3,000,000,000) of DNA, finding the mutations that cause diseases or traits is like finding a needle in a haystack.
For humans, the haystack gets more complicated! Some traits and diseases, such as height, depression, cancer, autism, and obesity, are outcomes of multiple variants interacting with each other and the environment. Then, there are also epigenetics and other factors that determine whether the mutations manifest themselves. Importantly, even though the SNPs reside in a gene, they may not change the gene’s genetic expression at all.
In fact, for depression, years of large-scale genetic association studies have failed to reproducibly identify genes that substantially increased the risk!3,4 It wasn’t until interactions with the environment and variability of symptoms were addressed in the study design that they started to find something.5
Analysis of twins and families indicates whether there are genetic components. Generally, if a trait runs in the family, it tends to have a genetic component.
Genome-wide association studies (GWAS) involve comparing the genomes of populations with and without the conditions of interest. Then, a statistical test is performed to identify SNPs and regions that are over-represented among people with the disease and absent in those without the disease.6
Many GWAS have been groundbreaking at identifying variants that contribute to the risk of type 2 diabetes, Parkinson’s disease, heart disorders, obesity, Crohn’s disease, and many types of cancers.
Most SNPs that are tested by direct-to-consumer tests have been identified in genome-wide association studies. These companies use the same technology, the SNP microarray, to identify your SNPs. This technology is relatively affordable and reliable, with a wholesale cost of approximately $40/sample.6 However, the tests are not without problems.
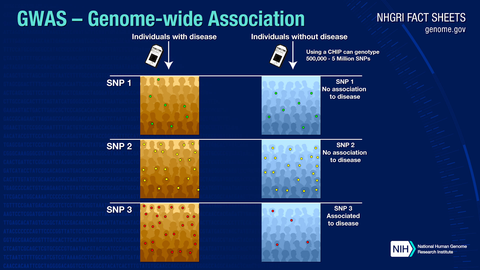
Image source:7 Caveats of Relying on Genetic-Based Health Advice Based on GWAS Data: Risk of Misinterpretation by the General Public
GWAS Do Not Prove Causality or Mechanism
GWAS “associates” a genetic variant with the risk of a trait. However, simply because umbrellas are associated with rain does not mean umbrellas cause rain. The SNP could just happen to be there.
Instead, finding an SNP inside of a gene being significantly associated with a trait means that you have a few reasonable hypotheses that may explain the association. Some SNPs change the encoded amino acids, while others don’t. You can test the hypotheses with biochemical experiments or by doing a genetic study in a model organism. In fact, a very small subset of SNPs tested by common SNP arrays has experimental proof of mechanism.
Notably, many SNPs do not affect the function of the genes in which they reside. The human genome is so complex that an SNP may influence the function of a gene 2 million base pairs away. The affected gene may not be the closest gene to the SNP.8 Without direct biochemical measurements, it is impossible to tell whether the SNPs increase, decrease, or do nothing to the gene function.
I’ve seen many genetic interpreter products infer SNP impacts based on the SNP’s location and what we know so far about the genes near the SNPs. While this could be considered “cutting-edge,” the interpretations are only as valid as a hypothesis and should be interpreted as such. Importantly, the lack of risk variants does not confirm that your risk is zero.
Now is a truly exciting era for genomic research, but we have barely scratched the surface of understanding how genes affect our health. Only a handful of well-known genes and their associated variants have been well-characterized enough to infer functional significance. Except for well-studied genes such as MTHFR, COMT, and ApoE, the data simply does not exist to explain the impact of these variants. In addition, most genetic software companies do not perform biochemical experiments to test gene function; they all rely on the same published literature. Therefore, for less-characterized genes, the genetic interpretations and health recommendations based on GWAS data are as good as guesswork.
As a result, if you send out your sample to different companies, you may find that the reports come back with opposite interpretations and recommendations on the same genotype!
Omission of Qualifying Information
GWAS typically provides qualifying information with respect to their findings.
As a hypothetical example, a genetic interpretation report may indicate that you have an SNP associated with being leaner if you consumed more monounsaturated fat.
The study may indicate that every tablespoon of monounsaturated fat consumed daily is associated with a 100g (approx. ¼ lb.) reduction in weight among obese participants. The association is “statistically significant.”
The first danger is when the association is communicated to lay consumers as “Monounsaturated fat helps you stay lean. Eat lots of monounsaturated fats.”
Is the statistically significant result practically significant? 100g of weight loss is barely practically significant for obese people. In real life, it’s typically far easier and better to lose 100g of fat any other way than taking olive oil shots. In fact, supplementing with olive oil shots will most likely have the opposite effect if you’re not changing your dietary habits otherwise.
Also, the weight loss tip from this study may not be applicable to any non-obese people or perhaps another ethnicity.
Thankfully, olive oil shots aren’t so deadly. However, when it comes to medications and medical interventions, the imprecision and omissions can be deadly.
Lack of Clinical Studies to Confirm the Safety and Effectiveness of Gene-Based Interventions
If nutrigenomics were to be perfectly scientific, every recommendation should be tested in clinical trials. The trials should include participants with and without the genotype of interest. The study design needs to also account for the placebo effect from the belief that they’re getting the best treatment for their genotype because the belief alone can confer health benefits. Alternatively, the study design could ensure that participants are completely unaware of their genetic and treatment/placebo status to eliminate such a placebo effect. Then, the outcomes for each group of participants should be statistically analyzed for significant effects.
Currently, most interpretations of SNP impacts and ways to address them are based on fairly loose scientific ground. On top of that, very few nutrigenomic clinical trials have successfully demonstrated that gene-based interventions are truly the best interventions.
A randomized controlled trial published in JAMA compared the effects of low-carb vs. low-fat on weight loss in 609 obese participants. They could not find significant effects based on genetic variants or baseline insulin secretion. It also turned out that both diets had comparable effects.9
Your SNPs May Not Matter All That Much
There are numerous ways your gene function can change other than SNPs. Much of these are influenced by epigenetics and the microbiome. You should consider that your genetics (SNPs + all kinds of variants) load the gun, while your environment pulls the trigger.
How Accurate are Genetic Tests Like 23andme, Ancestry, and Geno?
The companies claim that these technologies are 99% accurate. By this definition, if you’re determining hundreds of thousands of SNPs at the same time, you could estimate that at least around 1000s of SNPs (1%) could be incorrect from any given dataset. Therefore, these tests are not considered diagnostic, and the companies claim that you need to retest with a more authoritative lab.
The more problematic part is typically the interpretation of data and how to make sure that people understand it correctly. To date, the 23andme report is the only one that has been FDA-approved, since it only talks about well-studied variants. 23andme also had to test reading comprehension levels among lay readers to get the FDA approval.10
Privacy Issues With Testing Your Genes
There are worries about your personal health data being sold, stolen, or whether it can be used against you for insurance and employment purposes. Currently, there are laws against such practices.
Another problem is that testing your genes may reveal disease risks that you were not aware of and may not want to know. Importantly, it may reveal information about your family members or relatives without their consent. There have been a few cases where direct-to-consumer genetic tests revealed non-paternity or that the tester has been adopted.
Should You Test Your Genes?
My answer is that it is a very personal choice. Genetic testing and wading through the results are not for everyone.
I personally have tested my own genes and used genetic testing data as a health coach. However, I use the genetic data as hypothesis generators, not as answers. I also look at the studies behind the genes to find the qualifying information and confirm that the effect size is practically significant.
If you’re looking at your direct-to-consumer SNP data in 2020, you should exercise skepticism when you interpret the SNPs and health recommendations.
These tests and genetic reports are best for people with a biohacking or experimental mindset. They give you hypotheses of what could potentially work well for you, so you should test them, ideally in a placebo-controlled manner. MyJourney.science is a great resource for this. We are building out experimental templates and appropriate study designs for n = 1 experiments.
If you’re looking for answers and solid advice, work with a genetic counselor who understands the caveats and a health coach who can integrate all pieces of information to give you the solid advice you need.
References
https://www.pacb.com/blog/two-review-articles-assess-structural-variation-in-human-genomes/
Shastry, Barkur S. “SNPs: Impact on Gene Function and Phenotype.” Methods in Molecular Biology 578 (2009): 3–22. https://doi.org/10.1007/978-1-60327-411-1_1.
Border, Richard, Emma C. Johnson, Luke M. Evans, Andrew Smolen, Noah Berley, Patrick F. Sullivan, and Matthew C. Keller. “No Support for Historical Candidate Gene or Candidate Gene-by-Interaction Hypotheses for Major Depression Across Multiple Large Samples.” The American Journal of Psychiatry 176, no. 5 (2019): 376–87. https://doi.org/10.1176/appi.ajp.2018.18070881.
Lohoff, Faulk W. “Overview of the Genetics of Major Depressive Disorder.” Current Psychiatry Reports 12, no. 6 (2010): 539–546. https://europepmc.org/article/med/20848240.
Ormel, Johan, Catharina A. Hartman, and Harold Snieder. “The Genetics of Depression: Successful Genome-Wide Association Studies Introduce New Challenges.” Translational Psychiatry 9, no. 1 (2019): 114. https://doi.org/10.1038/s41398-019-0450-5.
Tam, Vivian, Nikunj Patel, Michelle Turcotte, Yohan Bossé, Guillaume Paré, and David Meyre. “Benefits and Limitations of Genome-Wide Association Studies.” Nature Reviews. Genetics 20, no. 8 (2019): 467–84. https://doi.org/10.1038/s41576-019-0127-1.
https://hbr.org/2020/09/23andmes-ceo-on-the-struggle-to-get-over-regulatory-hurdles.
Brodie, Aharon, Johnathan Roy Azaria, and Yanay Ofran. “How Far From the SNP May the Causative Genes Be?” Nucleic Acids Research 44, no. 13 (2016): 6046–54.
Gardner, Christopher D., John F. Trepanowski, Liana C. Del Gobbo, Michelle E. Hauser, Joseph Rigdon, John P. A. Ioannidis, Manisha Desai, and Abby C. King. 2018. “Effect of Low-Fat vs Low-Carbohydrate Diet on 12-Month Weight Loss in Overweight Adults and the Association With Genotype Pattern or Insulin Secretion: The DIETFITS Randomized Clinical Trial.” JAMA: The Journal of the American Medical Association 319, no. 7 (2018): 667–79. https://doi.org/10.1001/jama.2018.0245.
https://hbr.org/2020/09/23andmes-ceo-on-the-struggle-to-get-over-regulatory-hurdles.







